Endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes, however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like.[1] Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.[2]
Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence.[3] The nucleotide sequence recognized for cleavage by a restriction enzyme is called the restriction site. Typically, a restriction site will be a palindromic sequence about four to six nucleotides long. Most restriction endonucleases cleave the DNA strand unevenly, leaving complementary single-stranded ends. These ends can reconnect through hybridization and are termed "sticky ends". Once paired, the phosphodiester bonds of the fragments can be joined by DNA ligase. There are hundreds of restriction endonucleases known, each attacking a different restriction site. The DNA fragments cleaved by the same endonuclease can be joined together regardless of the origin of the DNA. Such DNA is called recombinant DNA; DNA formed by the joining of genes into new combinations.[4]Restriction endonucleases (restriction enzymes) are divided into three categories, Type I, Type II, and Type III, according to their mechanism of action. These enzymes are often used in genetic engineering to make recombinant DNA for introduction into bacterial, plant, or animal cells, as well as in synthetic biology.[5] One of the more famous endonucleases is Cas9.
Contents
1 Categories
2 Notations
3 Further discussion
4 DNA repair
5 Common endonucleases
6 Mutations
7 See also
8 References
Categories
Ultimately, there are three categories of restriction endonucleases that relatively contribute to the cleavage of specific sequences. The types I and III are large multisubunit complexes that include both the endonucleases and methylase activities. Type I can cleave at random sites of about 1000 base pairs or more from the recognition sequence and it requires ATP as source of energy. Type II behaves slightly differently and was first isolated by Hamilton Smith in 1970. They are simpler versions of the endonucleases and require no ATP in their degradation processes. Some examples of type II restriction endonucleases include BamHI, EcoRI, EcoRV, and Haelll. Type III, however, cleaves the DNA at about 25 base pairs from the recognition sequence and also requires ATP in the process.[4]
Notations
The commonly used notation for restriction endonucleases is of the form "vwxyz", where "vwx" names the life form (bacteria) where this restriction endonuclease may be found, "y" names the strain (and is optional), and "z" (in Roman numerals) indicates different restriction endonucleases in the same life form (bacteria). Thus for example, "EcoRI" means that the restriction endonuclease is found in Escherichia coli ("Eco"); strain RY13 ("R"), restriction endonuclease number "I". Another example: "HaeII" and "HaeIII" refer to bacterium Haemophilus aegyptius, number II and number III, respectively.[4]:64-64 The restriction enzymes used in molecular biology usually recognize short target sequences of about 4 – 8 base pairs. For instance, the EcoRI enzyme recognizes and cleaves the sequence 5' – GAATTC – 3'.[6]
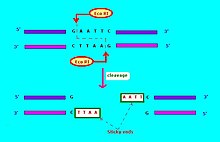
Restriction enzyme Eco RI
Restriction endonucleases come in several types. A restriction endonuclease typically requires a recognition site and a cleavage pattern (typically of nucleotide bases: A, C, G, T). If the recognition site is outside the region of the cleavage pattern, then the restriction endonuclease is referred to as Type I. If the recognition sequence overlaps with the cleavage sequence, then the restriction endonuclease restriction enzyme is Type II.
Further discussion
Restriction endonucleases may be found that cleave standard dsDNA (double-stranded DNA), or ssDNA (single-stranded DNA), or even RNA. This discussion is restricted to dsDNA; however, the discussion can be extended to the following:
- Standard dsDNA
- Non-standard DNA
- Holliday junctions
Triple-stranded DNA, quadruple-stranded DNA (G-quadruplex)- Double-stranded hybrids of DNA and RNA (one strand is DNA, the other strand is RNA)[4]:72–73
- Synthetic or artificial DNA (for example, containing bases other than A, C, G, T, refer to the work of Eric T. Kool). Research with synthetic codons, refer to the research by S. Benner, and enlarging the amino acid set in polypeptides, thus enlarging the proteome or proteomics, see the research by P. Schultz.[4]:chapter 3
In addition, research is now underway to construct synthetic or artificial restriction endonucleases, especially with recognition sites that are unique within a genome.
Restriction endonucleases or restriction enzymes typically cleave in two ways: blunt-ended or sticky-ended patterns. An example of a Type I restriction endonuclease.[4]:64
Furthermore, there exist DNA/RNA non-specific endonucleases, such as those that are found in Serratia marcescens, which act on dsDNA, ssDNA, and RNA.
DNA repair
Endonucleases play a role in DNA repair. AP endonuclease, specifically, catalyzes the incision of DNA exclusively at AP sites, and therefore prepares DNA for subsequent excision, repair synthesis and DNA ligation. For example, when depurination occurs, this lesion leaves a deoxyribose sugar with a missing base.[7] The AP endonuclease recognizes this sugar and essentially cuts the DNA at this site and then allows for DNA repair to continue.[8]E. coli cells contain two AP endonucleases: endonuclease IV (endoIV) and exonuclease III (exoIII) while in eukaryotes, there is only one AP endonuclease.[9]

Common endonucleases
Below are tables of common prokaryotic and eukaryotic endonucleases.[10]
| Prokaryotic Enzyme | Source | Comments |
|---|---|---|
| RecBCD enonuclease | E. coli | Partially ATP dependent; also an exonuclease; functions in recombination and repair |
| T7 endonuclease | phage T7 encoded (gene 3) | Essential for replication; preference for single stranded over double stranded DNA |
| T4 endonuclease IV | phage T4 encoded (denA) | Splits -TpC- sequence to yield 5'-dCMP- terminated oligonucleotides; chain length of product varies with conditions |
| Bal 31 endonuclease | Alteromonas espejiana | Also an exonuclease; nibbles away 3' and 5' ends of duplex DNA |
| EndonucleaseI (endo I) | E. coli (endA) | Periplasmic location; average chain length of product is 7; inhibited by tRNA; produces double stranded DNA break; produces nick when complexed with tRNA; endo I mutants grow normally |
| Micrococcal nuclease | Staphylococcus | Produces 3'-P termini; requires Ca2+; also acts on RNA; prefers single stranded DNA and AT-rich regions |
| Endonuclease II (endo VI, exo III) | E. coli (xth) | Cleavage next to AP site; also a 3'-->5' exonuclease; phosphomonoesterase on 3'-P termini |
| Eukaryotic Enzyme | Source | Comments |
|---|---|---|
| Neurospora endonuclease | Neurospora crassa | Also acts on RNA |
| S1-nuclease | Aspergillus oryzae | Also acts on RNA |
| P1-nuclease | Penicillium citrinum | Also acts on RNA |
| Mung bean nuclease I | mung bean sprouts | Also acts on RNA |
| Ustilago nuclease (Dnase I) | Ustilago maydis | Also acts on RNA |
| Dnase I | Bovine pancreas | Average chain length of product is 4; produces double strand break in presence of Mn2+ |
| AP endonuclease | Nucleus, mitochondria | Involved in DNA Base Excision Repair pathway |
| Endo R | HeLa cells | Specific for GC sites |
Mutations
Xeroderma pigmentosa is a rare, autosomal recessive disease caused by a defective UV-specific endonuclease. Patients with mutations are unable to repair DNA damage caused by sunlight.[11]
Sickle Cell anemia is a disease caused by a point mutation. The sequence altered by the mutation eliminates the recognition site for the restriction endonuclease MstII that recognizes the nucleotide sequence.[12]
tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Pontocerebellar hypoplasias (PCH) represent a group of neurodegenerative autosomal recessive disorders that is caused by mutations in three of the four different subunits of the tRNA-splicing endonuclease complex.[13]
See also
- Exonuclease
- Restriction endonuclease
- Nuclease
- Ribonuclease
- AP endonuclease
References
^ "Properties of Exonucleases and Endonucleases". New England BioLabs. 2017. Retrieved May 21, 2017..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output .citation q{quotes:"""""""'""'"}.mw-parser-output .citation .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-ws-icon a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-maint{display:none;color:#33aa33;margin-left:0.3em}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Slor, Hanoch (April 14, 1975). "Differenttation between exonucleases and endonucleases and between haplotomic and diplotomic endonucleases using 3H-DNA-coated wells of plastic depression plates as substrate". Nucleic Acids Research. 2: 897–903. doi:10.1093/nar/2.6.897.
^ Stephen T. Kilpatrick; Jocelyn E. Krebs; Lewin, Benjamin; Goldstein, Elliott (2011). Lewin's genes X. Boston: Jones and Bartlett. ISBN 0-7637-6632-1.
^ abcdef Cox M, Nelson DR, Lehninger AL (2005). Lehninger principles of biochemistry. San Francisco: W.H. Freeman. p. 952. ISBN 0-7167-4339-6.
^ Simon M (2010). Emergent computation: Emphasizing Bioinformatics. New York: Springer. p. 437. ISBN 1441919635.
^ Losick R, Watson JD, Baker TA, Bell S, Gann S, Levine MW (2008). Molecular biology of the gene. San Francisco: Pearson/Benjamin Cummings. ISBN 0-8053-9592-X.
^ Ellenberger T, Friedberg EC, Walker GS, Wolfram S, Wood RJ, Schultz R (2006). DNA repair and mutagenesis. Washington, D.C: ASM Press. ISBN 1-55581-319-4.
^ Alberts B (2002). Molecular biology of the cell. New York: Garland Science. ISBN 0-8153-3218-1.
^ Nishino T, Morikawa K (December 2002). "Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors". Oncogene. 21 (58): 9022–32. doi:10.1038/sj.onc.1206135. PMID 12483517.
^ Tania A. Baker; Kornberg, Arthur (2005). DNA replication. University Science. ISBN 1-891389-44-0.
^ Medical Biochemistry at a Glance. New York: Wiley. 2012. ISBN 0-470-65451-1.
^ Ferrier DR, Champe PC, Harvey RP (2008). Biochemistry. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins. ISBN 0-7817-6960-4.
^ Budde BS, Namavar Y, Barth PG, Poll-The BT, Nürnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, te Beek ET, Aronica E, van der Knaap MS, Höhne W, Toliat MR, Crow YJ, Steinling M, Voit T, Roelenso F, Brussel W, Brockmann K, Kyllerman M, Boltshauser E, Hammersen G, Willemsen M, Basel-Vanagaite L, Krägeloh-Mann I, de Vries LS, Sztriha L, Muntoni F, Ferrie CD, Battini R, Hennekam RC, Grillo E, Beemer FA, Stoets LM, Wollnik B, Nürnberg P, Baas F (September 2008). "tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia". Nat. Genet. 40 (9): 1113–8. doi:10.1038/ng.204. PMID 18711368.