Facultative aerobic organism
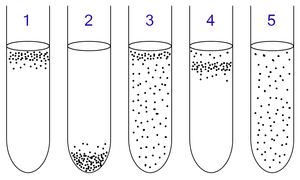
Aerobic and anaerobic bacteria can be identified by growing them in test tubes of thioglycolate broth:
1: Obligate aerobes need oxygen because they cannot ferment or respire anaerobically. They gather at the top of the tube where the oxygen concentration is highest.
2: Obligate anaerobes are poisoned by oxygen, so they gather at the bottom of the tube where the oxygen concentration is lowest.
3: Facultative anaerobes can grow with or without oxygen because they can metabolise energy aerobically or anaerobically. They gather mostly at the top because aerobic respiration generates more ATP than fermentation.
4: Microaerophiles need oxygen because they cannot ferment or respire anaerobically. However, they are poisoned by high concentrations of oxygen. They gather in the upper part of the test tube but not the very top.
5: Aerotolerant anaerobes do not require oxygen as they use fermentation to make ATP. Unlike obligate anaerobes, they are not poisoned by oxygen. They can be found evenly spread throughout the test tube.
A facultative anaerobe is an organism that makes ATP by aerobic respiration if oxygen is present, but is capable of switching to fermentation if oxygen is absent.
Some examples of facultatively anaerobic bacteria are Staphylococcus spp., Streptococcus spp.,[1]Escherichia coli, Salmonella, Listeria spp.[2] and Shewanella oneidensis. Certain eukaryotes are also facultative anaerobes, including fungi such as Saccharomyces cerevisiae[3] and many aquatic invertebrates such as Nereid (worm) polychaetes.[4]
See also
- Aerobic respiration
- Anaerobic respiration
- Fermentation
- Obligate aerobe
- Obligate anaerobe
- Microaerophile
References
^ Ryan KJ; Ray CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. pp. 261–271, 273–296. ISBN 0-8385-8529-9..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Singleton P (1999). Bacteria in Biology, Biotechnology and Medicine (5th ed.). Wiley. pp. 444–454. ISBN 0-471-98880-4.
^ Carlile MJ, Watkinson SC, Gooday GW (2001). The Fungi (2nd ed.). Academic Press. pp. 85–105. ISBN 0-12-738446-4.
^ Schöttler, U. (November 30, 1979). "On the Anaerobic Metabolism of Three Species of Nereis (Annelida)" (PDF). Marine Ecology Progress Series. 1: 249–54. doi:10.3354/meps001249. ISSN 1616-1599. Retrieved February 14, 2010.
External links
- Facultative Anaerobic Bacteria
- Obligate Anaerobic Bacteria
- Anaerobic Bacteria and Anaerobic Bacteria in the decomposition (stabilization) of organic matter.