Thiostrepton
 | |
| Names | |
|---|---|
| Other names Alaninamide, Bryamycin, Thiactin | |
| Identifiers | |
CAS Number |
|
3D model (JSmol) |
|
ChEMBL |
|
ECHA InfoCard | 100.014.304 |
PubChem CID |
|
InChI
| |
SMILES
| |
| Properties | |
Chemical formula | C72H85N19O18S5 |
Molar mass | 1664.83 g/mol |
| Appearance | White to off-white powder |
Melting point | 246 to 256 °C (475 to 493 °F; 519 to 529 K) |
Solubility in water | Insoluble |
Solubility in other solvents | Soluble in CHCl3, CH2Cl2, dioxane, pyridine, glacial acetic acid, DMF. Practically insoluble in the lower alcohols, nonpolar organic solvents, diluted aqueous acids or bases. May be dissolved by methanolic acid or base, but with decomposition.[2] |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Infobox references | |
Thiostrepton is a natural cyclic oligopeptide antibiotic of the thiopeptide class, derived from several strains of streptomycetes, such as Streptomyces azureus and Streptomyces laurentii. Thiostrepton is a natural product of the ribosomally synthesized and post-translationally modified peptide (RiPP) class.
Contents
1 History
2 Applications
3 Biosynthesis
3.1 Alternative mechanism for the formation of the dehydropiperidine core
4 Total synthesis
4.1 Building blocks
4.2 Synthetic scheme
5 References
History
Thiostrepton was discovered by Donovick et al. who described its antibacterial properties in 1955.[3]Dorothy Crowfoot Hodgkin solved the structure of thiostrepton in 1970.[4]
Early in 1978, Bycroft and Gowland[5]
proposed the biosynthesis of thiostrepton, which was still unclear until 2009. Several studies of thiopeptide biosynthesis[6][7][8][9] have been contemporarily published in 2009 and two of them (Liao et al. and Kelly et al.) included the similar biosynthesis of thiostrepton: it's ribosomally synthesized from thiostrepton biosynthetic genes (tsr genes) and posttranslational modification is needed.
A total synthesis of thiostrepton was completed by K.C. Nicolaou, et al. in 2004.[10][11]
Applications
Thiostrepton has been used in veterinary medicine in mastitis caused by gram-negative organisms and in dermatologic disorders. It is mostly used in complex ointments containing neomycin, nystatin, Thiostrepton and topical steroids. It is also active against gram-positive bacteria. It is notable that ointments for human usage contain neomycin, nystatin, and topical steroids, but no thiostrepton.
Thiostrepton was reported (in 2008) to exhibit activity against breast cancer cells through targeting the transcription factor forkhead box M1 (FOXM1),[12] also in 2011.[13]
It has also been shown to circumvent acquired cisplatin resistance in breast cancer cells under in invitro conditions.[14]
Thiostrepton is used in molecular biology as a reagent for both positive and negative selection of genes involved in nucleotide metabolism.
Biosynthesis
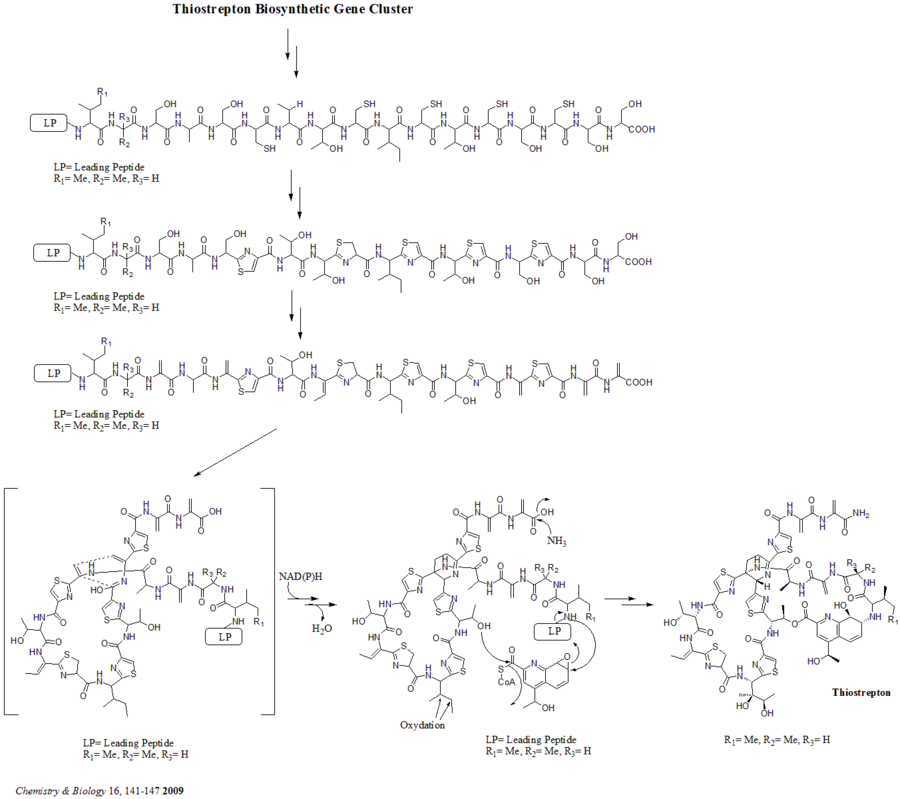
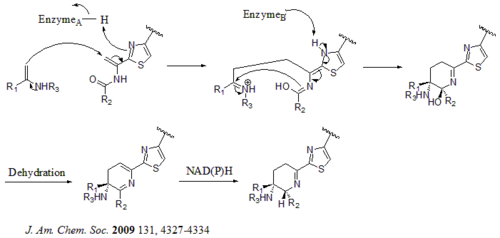
There are total 21 genes (tsrA~tsrU) in the biosynthetic gene cluster. The precursor of thiostrepton contains 58 amino acids in the peptide chain, which includes 41-aa leading peptide (LP) and 17-aa structural peptide (IASASCTTCICTCSCSS). Once the precursor is synthesized, cyclodehydratase tsrO and dehydrogenase tsrM catalyze the formation of thiazole or thiazoline from every cysteine residues in the peptide chain. After thiazole/thiazoline formation, dehydratases tsrJ, K and S then convert all the serine residues into dehydroalanines. A hetero Diels-Alder cyclization of the central dehydropiperidine (at S5, C13, and S14) has been suggested by Bycroft back to 1978 and been employed in the chemical synthesis of this core structure by Nicolaou et al. in 2005. An alternative mechanism of the dehydropiperidine formation has also been suggested by Kelly et al. in 2009. Nevertheless, based on experimental evidence, tsrN and L are suggested to be responsible for the hetero Diels-Alder cyclization. The quinaldic acid moiety is suggested to be synthesized by the nine genes tsrFAEBDUPQI from tryptophan and then results in the closure of quinaldic acid macrocycle. At last, tsrR serves as a candidate for the oxidation of the Ile residue to afford thiostrepton.
Alternative mechanism for the formation of the dehydropiperidine core
Total synthesis
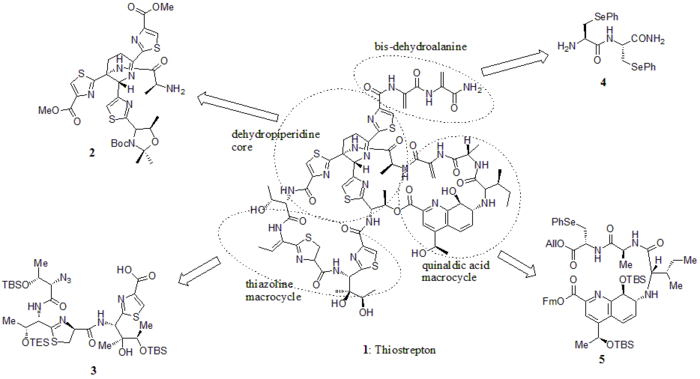
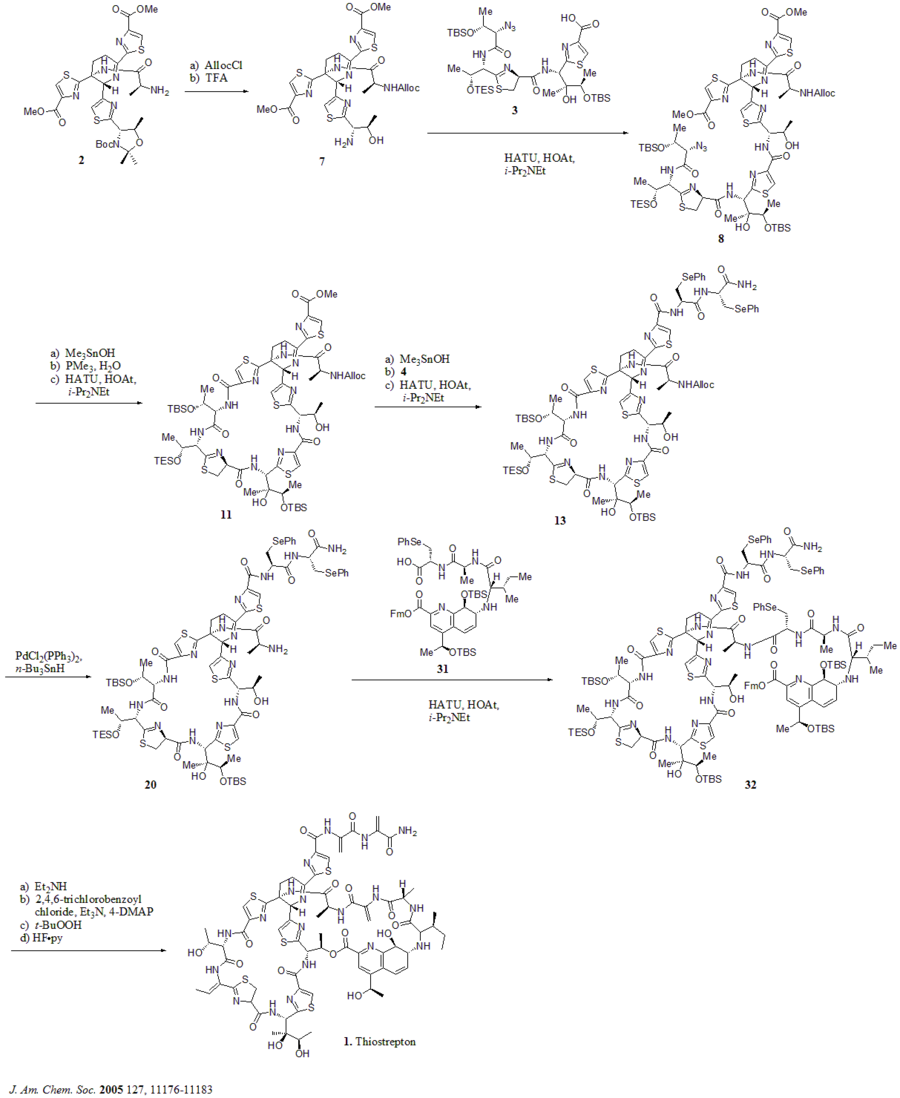
In 2005, Nicolaou et al. published the total synthesis of thiostrepton. At first, they constructed the key building blocks of thiostrepton (1): dehydropiperidine core (2), thiazoline macrocycle (3), bis-dehydroalanine tail (4), and quinaldic acid macrocycle (5). Then they assembled the building blocks sequentially as shown in the synthetic scheme (compound numbers are from the reference).
Building blocks
Synthetic scheme
References
^ Merck Index, 11th Edition, 9295.
^ Thiostrepton product page at Fermentek
^ Donovick R, Pagano JF, Stout HA, Weinstein MJ (1955). "Thiostrepton, a new antibiotic. I. In vitro studies". Antibiot Annu. 3: 554–9. PMID 13355325..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output .citation q{quotes:"""""""'""'"}.mw-parser-output .citation .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-ws-icon a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-maint{display:none;color:#33aa33;margin-left:0.3em}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Anderson B, Crowfoot Hodgkin D, Viswamitra MA (1970). "The Structure of Thiostrepton". Nature. 225 (5229): 223–235. doi:10.1038/225233a0.
^ Bycroft, Barrie W.; Gowland, Maxim S. (1978). "The structures of the highly modified peptide antibiotics micrococcin P1 and P2". Journal of the Chemical Society, Chemical Communications (6): 256. doi:10.1039/c39780000256. ISSN 0022-4936.
^ Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA (2009). "Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin". Proc. Natl. Acad. Sci. U.S.A. 106 (8): 2549–53. doi:10.1073/pnas.0900008106. PMC 2650375. PMID 19196969.
^ Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P (2009). "Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu". J. Am. Chem. Soc. 131 (16): 5946–55. doi:10.1021/ja900488a. PMID 19338336.
^ Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W (2009). "Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications". Chem. Biol. 16 (2): 141–7. doi:10.1016/j.chembiol.2009.01.007. PMC 2676563. PMID 19246004.
^ Kelly WL, Pan L, Li C (2009). "Thiostrepton biosynthesis: prototype for a new family of bacteriocins". J. Am. Chem. Soc. 131 (12): 4327–34. doi:10.1021/ja807890a. PMID 19265401.
^ Nicolaou, K. C.; Zak, Mark; Safina, Brian S.; Estrada, Anthony A.; Lee, Sang Hyup; Nevalainen, Marta (2005). "Total Synthesis of Thiostrepton. Assembly of Key Building Blocks and Completion of the Synthesis". Journal of the American Chemical Society. 127 (31): 11176–11183. doi:10.1021/ja052934z. ISSN 0002-7863.
^ Nicolaou KC, Safina BS, Zak M, Lee SH, Nevalainen M, Bella M, Estrada AA, Funke C, Zécri FJ, Bulat S (2005). "Total synthesis of thiostrepton. Retrosynthetic analysis and construction of key building blocks". J. Am. Chem. Soc. 127 (31): 11159–75. doi:10.1021/ja0529337. PMID 16076224.
^ Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW (July 2008). "Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression". Mol. Cancer Ther. 7 (7): 2022–32. doi:10.1158/1535-7163.MCT-08-0188. PMID 18645012.
^ http://www.news-medical.net/news/20110822/Scientists-reveal-how-thiostrepton-blocks-FOXM1-protein-prevents-breast-cancer-development.aspx Scientists reveal how thiostrepton blocks FOXM1 protein, prevents breast cancer development. 2011
^ Kwok JM; Peck B; Monteiro LJ; Schwenen HD; Millour J; Coombes RC; Myatt SS; Lam EW. (January 2010). "FOXM1 confers acquired cisplatin resistance in breast cancer cells". Molecular Cancer Research. 8 (1): 24–34. doi:10.1158/1541-7786.MCR-09-0432. PMC 2809047. PMID 20068070.